Computational Systems Biology
The group’s research aims to understand the function and dysfunctions of biological systems on a molecular level, taking into account the complexity arising from the interplay of biomolecules in large regulatory networks. This work is enabled by the availability of big biological datasets that are generated by high-throughput, biochemical profiling methods. Relevant data include transcriptomics, proteomics, or epigenomics, which are often assayed in combination (“multi-omics”) and at cellular resolution (“single-cell”). The computational methods we use mirror this biological complexity, spanning advanced statistics, machine learning, and network science.
Key contributions
JAK-STAT signaling maintains homeostasis in T cells and macrophages
Fortelny*, Farlik*, et al., Nature Immunology (2024)
https://doi.org/10.1038/s41590-024-01804-1
Systematic functional screening of chromatin factors identifies strong lineage and disease dependencies in normal and malignant haematopoiesis
Lara-Astiaso*, Goñi-Salaverri*, Mendieta-Esteban* et al., Nature Genetics (2023)
https://doi.org/10.1038/s41588-023-01471-2
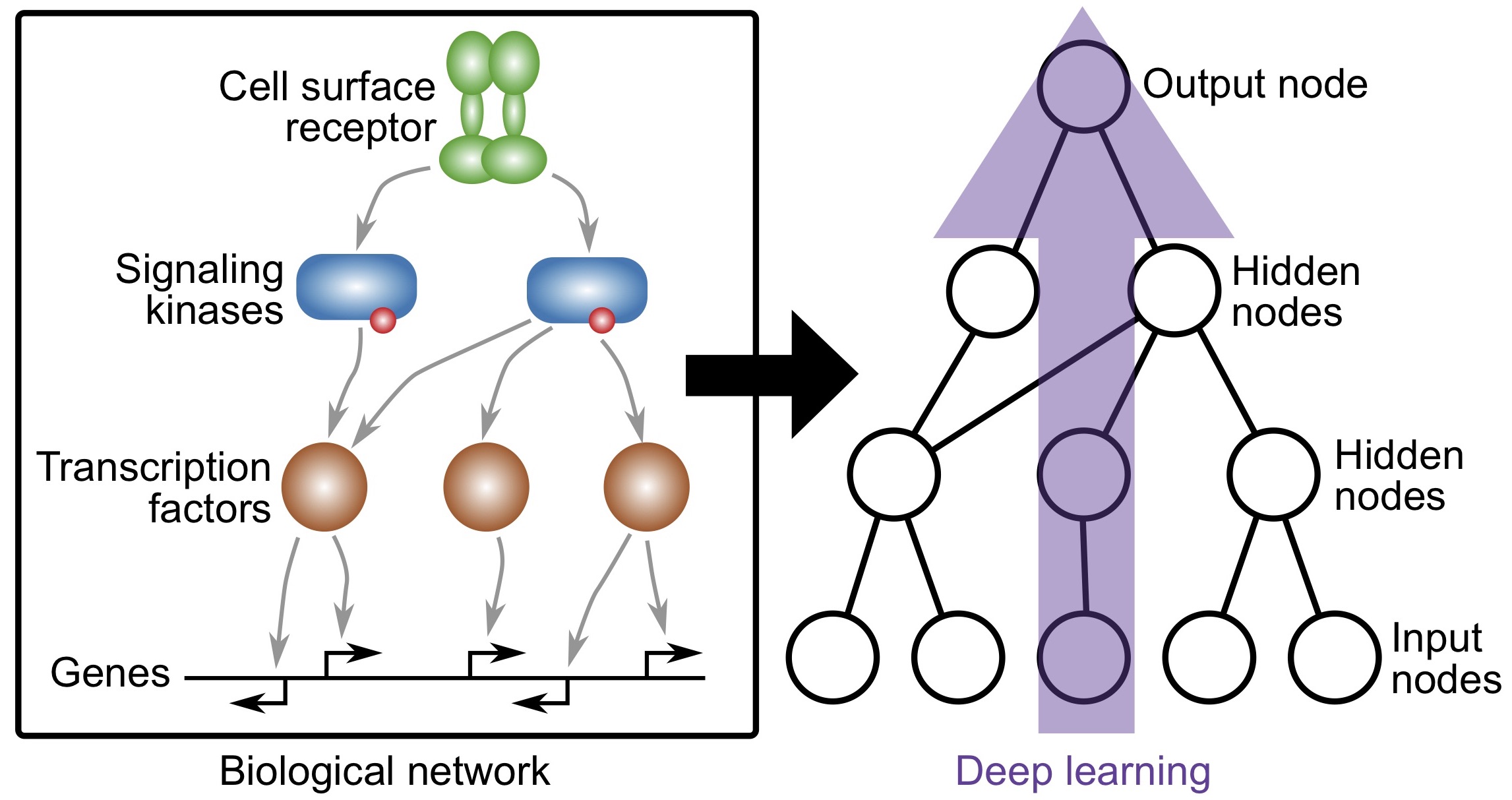
Knowledge-primed neural networks enable biologically interpretable deep learning on single-cell sequencing data
Fortelny and Bock, Genome Biology (2020)
https://doi.org/10.1186/s13059-020-02100-5
Structural cells are key regulators of organ-specific immune responses
Krausgruber* and Fortelny* et al., Nature (2020)
https://doi.org/10.1038/s41586-020-2424-4
Can we predict protein from mRNA levels?
Fortelny et al., Nature [BCA] (2017)
https://doi.org/10.1038/nature22293




