Home | Contact | Positions | Publications | Links
Structure and function of proprotein convertases
Many secretory and membrane-bound proteins are synthesized as precursor proteins within the cell (Fig. 1a). These so-called proproteins must first be activated to gain biological activity. This process is often regulated by limited proteolysis which includes the cleavage of a specific portion by a protease from the target protein. Alternatively, enzymes can also be inactivated through limited proteolysis. Alongside glycosylation, phosphorylation, and other chemical modifications, limited proteolysis is an example of post-translational modifications of proteins. The proprotein convertases (PCs), a group of serine proteases, play a prominent role in the proteolytic activation of many proproteins.
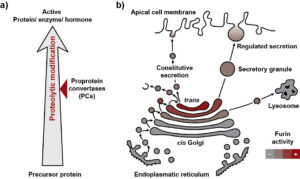
Figure 1 Physiological function of proprotein convertases.
Mammals possess nine different representatives of PCs, which are characterized by stringent sequence specificity. These proteases typically recognize multiple amino acids within the recognition motif of their substrates. Furin can be considered as a prototypical representative of a subgroup of PCs that cleave substrates C-terminal to poly-basic recognition sequences of the type R-X-(K/R)-R↓ (X represents any amino acid, K/R represents lysine or arginine at that position, “↓” symbolizes the scisile peptide bond). The high sequence specificity of PCs ensures that only the target proteins are cleaved, preventing uncontrolled proteolytic degradation of other proteins within the secretory apparatus.
Furthermore, it is crucial for the cell to prevent enzymes, receptors, or growth factors from being activated at the wrong location and/or at the wrong time. Therefore, there are different mechanisms to regulate the activity of the proprotein convertases (PCs). On one hand, PCs are transported to specific compartments of the secretory apparatus. On the other hand, the PCs themselves are synthesized as inactive proproteins and undergo auto-activation at their target compartment. This process involves a series of auto-proteolytic cleavages within the prodomain. The prodomain serves as a sensor that triggers auto-activation once a more or less acidic pH is present in the target compartment. For example, Furin is activated at pH 6.0 in the late Golgi (Fig. 1b). Through transport processes, Furin also reaches secretory vesicles and the cell surface and can be recycled again to the late Golgi.
Structural basis of proprotein convertase activation
As part of a project funded by the Austrian Science Fund (FWF) ( Project P 36648), we are investigating the mechanisms of auto-activation of proprotein convertases (PCs). We are particularly interested in the structural changes of the proteases that occur during the activation process. Additionally, PCs differ in their activation profiles. Within this project, we aim to further examine the differences in the activation mechanisms of different PC family members. Open positions on the project are announced here.
Mechanisms of substrate recognition and catalysis
Compared to other proteases, PCs exhibit particularly high substrate specificity. In this regard, the proteases recognize multiple positively charged amino acids within the recognition motif of their substrates. However, proteins and peptides with single arginines or lysines are not cleaved by PCs. We are interested in studying the interactions between the amino acids of the poly-basic recognition motif and the substrate-binding pocket, which form the basis of the substrate selectivity (Fig. 2). Detailed knowledge of these mechanisms is also valuable for structure-based development of improved inhibitors (see below).
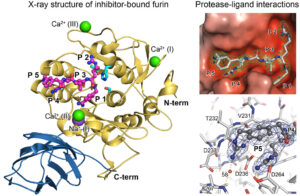
Figure 2 Crystal structure of furin bount to a substrate-like inhibitor and protease-inhibitor interactions.
Structural investigations have shown that the substrate-binding pocket of Furin possesses high plasticity and can exist in several specific conformations. For instance, we have shown that ligand binding induces a conformational change in the substrate-binding pocket from the so-called “OFF-state” to the “ON-state.” This conformational change also affects the state of the active site and other structural regions of Furin.
Another specific structural change in the substrate-binding pocket of Furin is triggered by the binding of dichlorophenylpyridine-based inhibitors (Fig. 3). In this case, the chlorinated phenyl moiety displaces a tryptophan side chain and binds to a so-called cryptic binding pocket. Interestingly, similar structural properties are observed distant to the substrate-binding pocket, which are also found during the transition from the “OFF-state” to the “ON-state.”
We are particularly interested in the conformational dynamics of Furin and molecular switches that regulate the catalytic activity of Furin.

Figure 3 Crystal structure of furin in complex with a dichlorophenylpyridine-based inhibitor.
One drug against many diseases – development of Furin inhibitors.
Furin is a promising target for the development of drugs against different diseases. In collaboration with chemists and virologists, we are developing inhibitors for the treatment of infectious diseases.




